Introduction
These are the results for the 2008 running of the Audio Genre Classification task. For background information about this task set please refer to the 2008:Audio Genre Classification page.
General Legend
Team ID
CL1 = C. Cao, M. Li 1
CL2 = C. Cao, M. Li 2
GP1 = G. Peeters
GT1 (mono) = G. Tzanetakis
GT2 (stereo) = G. Tzanetakis
GT3 (multicore) = G. Tzanetakis
LRPPI1 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 1
LRPPI2 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 2
LRPPI3 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 3
LRPPI4 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 4
ME1 = I. M. Mandel, D. P. W. Ellis 1
ME2 = I. M. Mandel, D. P. W. Ellis 2
ME3 = I. M. Mandel, D. P. W. Ellis 3
Overall Summary Results
Task 1 (MIXED) Results
MIREX 2008 Audio Genre Classification Summary Results - Raw Classification Accuracy Averaged Over Three Train/Test Folds
| Participant |
Average Classifcation Accuracy |
| CL1 |
62.04% |
| CL2 |
63.39% |
| GP1 |
63.90% |
| GT1 |
64.71% |
| GT2 |
66.41% |
| GT3 |
65.62% |
| LRPPI1 |
65.06% |
| LRPPI2 |
62.26% |
| LRPPI3 |
60.84% |
| LRPPI4 |
60.46% |
| ME1 |
65.41% |
| ME2 |
65.30% |
| ME3 |
65.20% |
download these results as csv
Accuracy Across Folds
| Classification fold |
CL1 |
CL2 |
GP1 |
GT1 |
GT2 |
GT3 |
LRPPI1 |
LRPPI2 |
LRPPI3 |
LRPPI4 |
ME1 |
ME2 |
ME3 |
| 0 |
0.592 |
0.598 |
0.634 |
0.639 |
0.642 |
0.654 |
0.650 |
0.610 |
0.598 |
0.606 |
0.631 |
0.631 |
0.628 |
| 1 |
0.644 |
0.661 |
0.634 |
0.651 |
0.682 |
0.664 |
0.669 |
0.637 |
0.626 |
0.617 |
0.668 |
0.665 |
0.666 |
| 2 |
0.625 |
0.643 |
0.649 |
0.652 |
0.669 |
0.651 |
0.633 |
0.622 |
0.602 |
0.592 |
0.663 |
0.662 |
0.663 |
download these results as csv
Accuracy Across Categories
| Class |
CL1 |
CL2 |
GP1 |
GT1 |
GT2 |
GT3 |
LRPPI1 |
LRPPI2 |
LRPPI3 |
LRPPI4 |
ME1 |
ME2 |
ME3 |
| BAROQUE |
0.616 |
0.637 |
0.750 |
0.669 |
0.724 |
0.673 |
0.673 |
0.660 |
0.666 |
0.629 |
0.754 |
0.759 |
0.757 |
| BLUES |
0.711 |
0.741 |
0.674 |
0.690 |
0.677 |
0.701 |
0.700 |
0.703 |
0.713 |
0.689 |
0.713 |
0.706 |
0.706 |
| CLASSICAL |
0.608 |
0.598 |
0.592 |
0.559 |
0.649 |
0.606 |
0.563 |
0.603 |
0.559 |
0.524 |
0.666 |
0.669 |
0.672 |
| COUNTRY |
0.624 |
0.596 |
0.697 |
0.793 |
0.830 |
0.679 |
0.669 |
0.640 |
0.621 |
0.617 |
0.660 |
0.656 |
0.653 |
| EDANCE |
0.560 |
0.591 |
0.536 |
0.590 |
0.624 |
0.648 |
0.672 |
0.626 |
0.646 |
0.686 |
0.657 |
0.649 |
0.639 |
| JAZZ |
0.679 |
0.699 |
0.606 |
0.627 |
0.682 |
0.626 |
0.640 |
0.602 |
0.566 |
0.574 |
0.679 |
0.676 |
0.680 |
| METAL |
0.677 |
0.709 |
0.750 |
0.713 |
0.656 |
0.733 |
0.707 |
0.642 |
0.623 |
0.643 |
0.612 |
0.627 |
0.629 |
| RAPHIPHOP |
0.809 |
0.823 |
0.873 |
0.846 |
0.846 |
0.854 |
0.860 |
0.837 |
0.826 |
0.848 |
0.841 |
0.836 |
0.837 |
| ROCKROLL |
0.420 |
0.418 |
0.406 |
0.384 |
0.414 |
0.447 |
0.448 |
0.391 |
0.377 |
0.391 |
0.450 |
0.450 |
0.448 |
| ROMANTIC |
0.501 |
0.527 |
0.508 |
0.602 |
0.540 |
0.597 |
0.574 |
0.523 |
0.488 |
0.444 |
0.510 |
0.505 |
0.500 |
download these results as csv
MIREX 2008 Audio Genre Classification Evaluation Logs and Confusion Matrices
MIREX 2008 Audio Genre Classification Run Times
| Participant |
Runtime (hh:mm) / Fold |
| CL1 |
Feat Ex: 01:29 Train/Classify: 0:33 |
| CL2 |
Feat Ex: 01:31 Train/Classify: 01:01 |
| GP1 |
Feat Ex: 11:37 Train/Classify: 00:25 |
| GT1 |
Feat Ex/Train/Classify: 00:36 |
| GT2 |
Feat Ex/Train/Classify: 00:35 |
| GT3 |
Feat Ex: 00:12 Train/Classify: 00:01 |
| LRPPI1 |
Feat Ex: 28:50 Train/Classify: 00:02 |
| LRPPI2 |
Feat Ex: 28:50 Train/Classify: 00:17 |
| LRPPI3 |
Feat Ex: 28:50 Train/Classify: 00:20 |
| LRPPI4 |
Feat Ex: 28:50 Train/Classify: 00:35 |
| ME1 |
Feat Ex: 3:35 Train/Classify: 00:02 |
| ME2 |
Feat Ex: 3:35 Train/Classify: 00:02 |
| ME3 |
Feat Ex: 3:35 Train/Classify: 00:02 |
download these results as csv
CSV Files Without Rounding
audiogenre_results_fold.csv
audiogenre_results_class.csv
Results By Algorithm
(.tar.gz)
CL1 = C. Cao, M. Li 1
CL2 = C. Cao, M. Li 2
LRPPI1 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 1
LRPPI2 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 2
LRPPI3 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 3
LRPPI4 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 4
ME1 = I. M. Mandel, D. P. W. Ellis 1
ME2 = I. M. Mandel, D. P. W. Ellis 2
ME3 = I. M. Mandel, D. P. W. Ellis 3
GP = G. Peeters
GT1 = G. Tzanetakis
GT2 = G. Tzanetakis
GT3 = G. Tzanetakis
Task 2 (LATIN) Results
MIREX 2008 Audio Genre Classification Summary Results - Raw Classification Accuracy Averaged Over Three Train/Test Folds
| Participant |
Average Classifcation Accuracy |
| CL1 |
65.17% |
| CL2 |
64.04% |
| GP1 |
62.72% |
| GT1 |
53.65% |
| GT2 |
53.79% |
| GT3 |
53.67% |
| LRPPI1 |
58.64% |
| LRPPI2 |
62.23% |
| LRPPI3 |
59.55% |
| LRPPI4 |
59.00% |
| ME1 |
54.15% |
| ME2 |
54.70% |
| ME3 |
54.99% |
download these results as csv
Accuracy Across Folds
| Classification fold |
CL1 |
CL2 |
GP1 |
GT1 |
GT2 |
GT3 |
LRPPI1 |
LRPPI2 |
LRPPI3 |
LRPPI4 |
ME1 |
ME2 |
ME3 |
| 0 |
0.755 |
0.750 |
0.694 |
0.674 |
0.677 |
0.657 |
0.661 |
0.697 |
0.671 |
0.680 |
0.681 |
0.684 |
0.685 |
| 1 |
0.541 |
0.528 |
0.553 |
0.435 |
0.435 |
0.422 |
0.512 |
0.573 |
0.526 |
0.506 |
0.403 |
0.409 |
0.415 |
| 2 |
0.660 |
0.644 |
0.634 |
0.501 |
0.502 |
0.531 |
0.587 |
0.597 |
0.590 |
0.585 |
0.541 |
0.548 |
0.550 |
download these results as csv
Accuracy Across Categories
| Class |
CL1 |
CL2 |
GP1 |
GT1 |
GT2 |
GT3 |
LRPPI1 |
LRPPI2 |
LRPPI3 |
LRPPI4 |
ME1 |
ME2 |
ME3 |
| axe |
0.753 |
0.745 |
0.558 |
0.637 |
0.640 |
0.695 |
0.529 |
0.560 |
0.537 |
0.528 |
0.679 |
0.679 |
0.681 |
| bachata |
0.957 |
0.622 |
0.969 |
0.595 |
0.592 |
0.587 |
0.957 |
0.950 |
0.956 |
0.957 |
0.932 |
0.935 |
0.935 |
| bolero |
0.630 |
0.633 |
0.768 |
0.702 |
0.705 |
0.746 |
0.683 |
0.726 |
0.646 |
0.668 |
0.664 |
0.662 |
0.666 |
| forro |
0.356 |
0.335 |
0.270 |
0.146 |
0.145 |
0.127 |
0.258 |
0.342 |
0.292 |
0.287 |
0.174 |
0.186 |
0.188 |
| gaucha |
0.501 |
0.491 |
0.345 |
0.348 |
0.348 |
0.299 |
0.345 |
0.357 |
0.327 |
0.338 |
0.435 |
0.436 |
0.434 |
| merengue |
0.895 |
0.898 |
0.897 |
0.812 |
0.806 |
0.784 |
0.847 |
0.794 |
0.825 |
0.833 |
0.698 |
0.699 |
0.728 |
| pagode |
0.355 |
0.368 |
0.303 |
0.307 |
0.297 |
0.249 |
0.322 |
0.391 |
0.361 |
0.318 |
0.231 |
0.240 |
0.243 |
| salsa |
0.886 |
0.850 |
0.750 |
0.715 |
0.719 |
0.668 |
0.788 |
0.793 |
0.769 |
0.766 |
0.698 |
0.710 |
0.716 |
| sertaneja |
0.209 |
0.205 |
0.200 |
0.159 |
0.186 |
0.212 |
0.132 |
0.227 |
0.210 |
0.170 |
0.090 |
0.090 |
0.094 |
| tango |
0.590 |
0.587 |
0.585 |
0.588 |
0.588 |
0.582 |
0.592 |
0.581 |
0.586 |
0.590 |
0.588 |
0.588 |
0.588 |
download these results as csv
MIREX 2008 Audio Genre Classification Evaluation Logs and Confusion Matrices
MIREX 2008 Audio Genre Classification Run Times
| Participant |
Runtime (hh:mm) / Fold |
| CL1 |
Feat Ex: 00:47 Train/Classify: 0:13 |
| CL2 |
Feat Ex: 00:48 Train/Classify: 00:23 |
| GP1 |
Feat Ex: 07:12 Train/Classify: 00:15 |
| GT1 |
Feat Ex/Train/Classify: 00:16 |
| GT2 |
Feat Ex/Train/Classify: 00:17 |
| GT3 |
Feat Ex: 00:06 Train/Classify: 00:00 (6 sec) |
| LRPPI1 |
Feat Ex: 15:33 Train/Classify: 00:01 |
| LRPPI2 |
Feat Ex: 15:33 Train/Classify: 00:06 |
| LRPPI3 |
Feat Ex: 15:33 Train/Classify: 00:06 |
| LRPPI4 |
Feat Ex: 15:33 Train/Classify: 00:11 |
| ME1 |
Feat Ex: 1:58 Train/Classify: 00:00 (28 sec) |
| ME2 |
Feat Ex: 1:58 Train/Classify: 00:00 (28 sec) |
| ME3 |
Feat Ex: 1:58 Train/Classify: 00:00 (28 sec) |
download these results as csv
CSV Files Without Rounding
audiolatin_results_fold.csv
audiolatin_results_class.csv
Results By Algorithm
(.tar.gz)
CL1 = C. Cao, M. Li 1
CL2 = C. Cao, M. Li 2
GP1 = G. Peeters
GT1 = G. Tzanetakis
GT2 = G. Tzanetakis
GT3 = G. Tzanetakis
LRPPI1 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 1
LRPPI2 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 2
LRPPI3 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 3
LRPPI4 = T. Lidy, A. Rauber, A. Pertusa, P. Peonce de Leon, J. M. I├▒esta 4
ME1 = I. M. Mandel, D. P. W. Ellis 1
ME2 = I. M. Mandel, D. P. W. Ellis 2
ME3 = I. M. Mandel, D. P. W. Ellis 3
Friedman's Test for Significant Differences
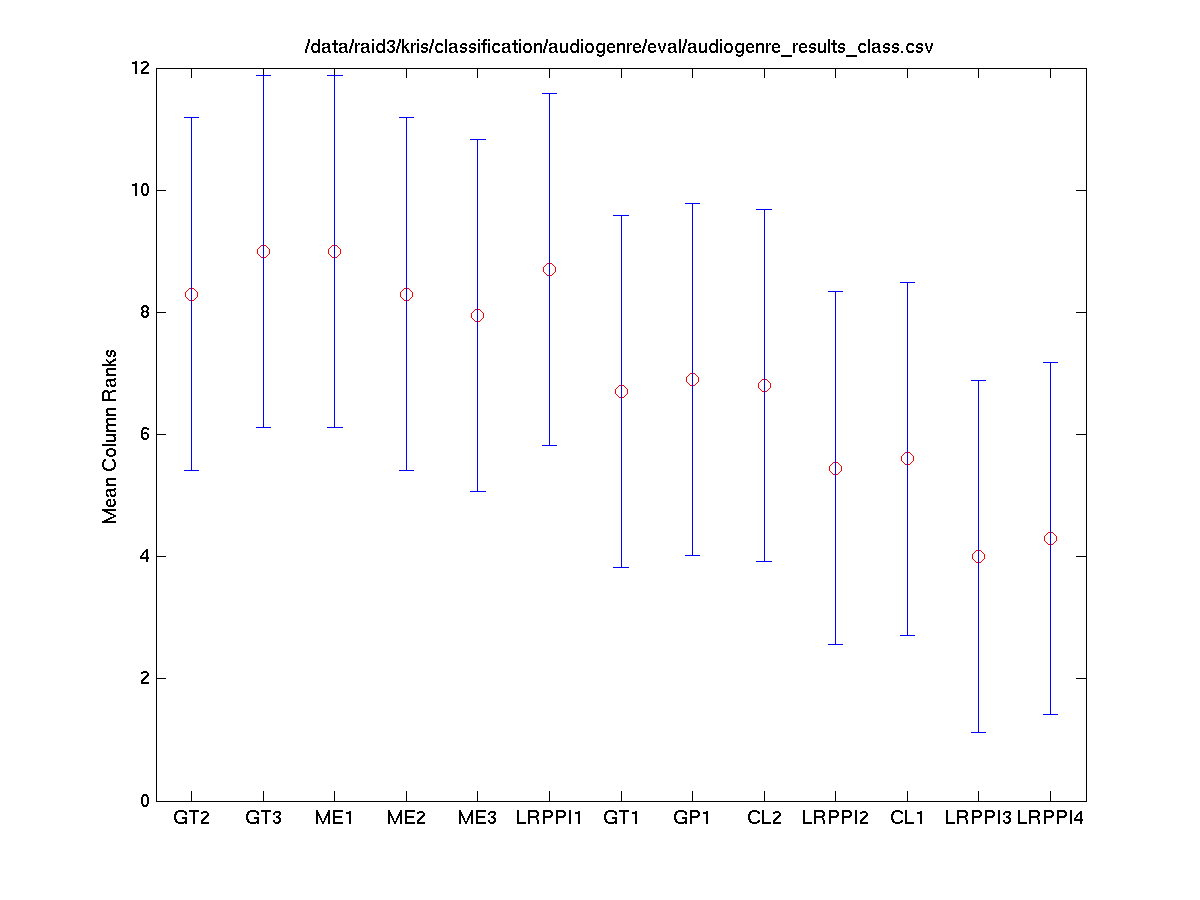
Task 1 (Mixed) Classes vs. Systems
The Friedman test was run in MATLAB against the average accuracy for each class.
Friedman's Anova Table
| Source |
SS |
df |
MS |
Chi-sq |
Prob>Chi-sq |
| Columns |
243.6 |
10 |
24.36 |
22.15 |
0.0144 |
| Error |
856.4 |
90 |
9.5156 |
|
|
| Total |
1100 |
109 |
|
|
|
download these results as csv
Tukey-Kramer HSD Multi-Comparison
The Tukey-Kramer HSD multi-comparison data below was generated using the following MATLAB instruction.
Command: [c, m, h, gnames] = multicompare(stats, 'ctype', 'tukey-kramer', 'estimate', 'friedman', 'alpha', 0.05);
| TeamID |
TeamID |
Lowerbound |
Mean |
Upperbound |
Significance |
| CL1 |
CL2 |
-5.5740 |
-0.8000 |
3.9740 |
FALSE |
| CL1 |
GP1 |
-5.1740 |
-0.4000 |
4.3740 |
FALSE |
| CL1 |
GT1 |
-5.0740 |
-0.3000 |
4.4740 |
FALSE |
| CL1 |
GT2 |
-3.3740 |
1.4000 |
6.1740 |
FALSE |
| CL1 |
GT3 |
-3.5740 |
1.2000 |
5.9740 |
FALSE |
| CL1 |
LRPPI1 |
-3.4740 |
1.3000 |
6.0740 |
FALSE |
| CL1 |
LRPPI2 |
-2.3740 |
2.4000 |
7.1740 |
FALSE |
| CL1 |
LRPPI3 |
-2.4740 |
2.3000 |
7.0740 |
FALSE |
| CL1 |
LRPPI4 |
-1.1740 |
3.6000 |
8.3740 |
FALSE |
| CL1 |
ME1 |
-1.1740 |
3.6000 |
8.3740 |
FALSE |
| CL2 |
GP1 |
-4.3740 |
0.4000 |
5.1740 |
FALSE |
| CL2 |
GT1 |
-4.2740 |
0.5000 |
5.2740 |
FALSE |
| CL2 |
GT2 |
-2.5740 |
2.2000 |
6.9740 |
FALSE |
| CL2 |
GT3 |
-2.7740 |
2.0000 |
6.7740 |
FALSE |
| CL2 |
LRPPI1 |
-2.6740 |
2.1000 |
6.8740 |
FALSE |
| CL2 |
LRPPI2 |
-1.5740 |
3.2000 |
7.9740 |
FALSE |
| CL2 |
LRPPI3 |
-1.6740 |
3.1000 |
7.8740 |
FALSE |
| CL2 |
LRPPI4 |
-0.3740 |
4.4000 |
9.1740 |
FALSE |
| CL2 |
ME1 |
-0.3740 |
4.4000 |
9.1740 |
FALSE |
| GP1 |
GT1 |
-4.6740 |
0.1000 |
4.8740 |
FALSE |
| GP1 |
GT2 |
-2.9740 |
1.8000 |
6.5740 |
FALSE |
| GP1 |
GT3 |
-3.1740 |
1.6000 |
6.3740 |
FALSE |
| GP1 |
LRPPI1 |
-3.0740 |
1.7000 |
6.4740 |
FALSE |
| GP1 |
LRPPI2 |
-1.9740 |
2.8000 |
7.5740 |
FALSE |
| GP1 |
LRPPI3 |
-2.0740 |
2.7000 |
7.4740 |
FALSE |
| GP1 |
LRPPI4 |
-0.7740 |
4.0000 |
8.7740 |
FALSE |
| GP1 |
ME1 |
-0.7740 |
4.0000 |
8.7740 |
FALSE |
| GT1 |
GT2 |
-3.0740 |
1.7000 |
6.4740 |
FALSE |
| GT1 |
GT3 |
-3.2740 |
1.5000 |
6.2740 |
FALSE |
| GT1 |
LRPPI1 |
-3.1740 |
1.6000 |
6.3740 |
FALSE |
| GT1 |
LRPPI2 |
-2.0740 |
2.7000 |
7.4740 |
FALSE |
| GT1 |
LRPPI3 |
-2.1740 |
2.6000 |
7.3740 |
FALSE |
| GT1 |
LRPPI4 |
-0.8740 |
3.9000 |
8.6740 |
FALSE |
| GT1 |
ME1 |
-0.8740 |
3.9000 |
8.6740 |
FALSE |
| GT2 |
GT3 |
-4.9740 |
-0.2000 |
4.5740 |
FALSE |
| GT2 |
LRPPI1 |
-4.8740 |
-0.1000 |
4.6740 |
FALSE |
| GT2 |
LRPPI2 |
-3.7740 |
1.0000 |
5.7740 |
FALSE |
| GT2 |
LRPPI3 |
-3.8740 |
0.9000 |
5.6740 |
FALSE |
| GT2 |
LRPPI4 |
-2.5740 |
2.2000 |
6.9740 |
FALSE |
| GT2 |
ME1 |
-2.5740 |
2.2000 |
6.9740 |
FALSE |
| GT3 |
LRPPI1 |
-4.6740 |
0.1000 |
4.8740 |
FALSE |
| GT3 |
LRPPI2 |
-3.5740 |
1.2000 |
5.9740 |
FALSE |
| GT3 |
LRPPI3 |
-3.6740 |
1.1000 |
5.8740 |
FALSE |
| GT3 |
LRPPI4 |
-2.3740 |
2.4000 |
7.1740 |
FALSE |
| GT3 |
ME1 |
-2.3740 |
2.4000 |
7.1740 |
FALSE |
| LRPPI1 |
LRPPI2 |
-3.6740 |
1.1000 |
5.8740 |
FALSE |
| LRPPI1 |
LRPPI3 |
-3.7740 |
1.0000 |
5.7740 |
FALSE |
| LRPPI1 |
LRPPI4 |
-2.4740 |
2.3000 |
7.0740 |
FALSE |
| LRPPI1 |
ME1 |
-2.4740 |
2.3000 |
7.0740 |
FALSE |
| LRPPI2 |
LRPPI3 |
-4.8740 |
-0.1000 |
4.6740 |
FALSE |
| LRPPI2 |
LRPPI4 |
-3.5740 |
1.2000 |
5.9740 |
FALSE |
| LRPPI2 |
ME1 |
-3.5740 |
1.2000 |
5.9740 |
FALSE |
| LRPPI3 |
LRPPI4 |
-3.4740 |
1.3000 |
6.0740 |
FALSE |
| LRPPI3 |
ME1 |
-3.4740 |
1.3000 |
6.0740 |
FALSE |
| LRPPI4 |
ME1 |
-4.7740 |
0.0000 |
4.7740 |
FALSE |
download these results as csv
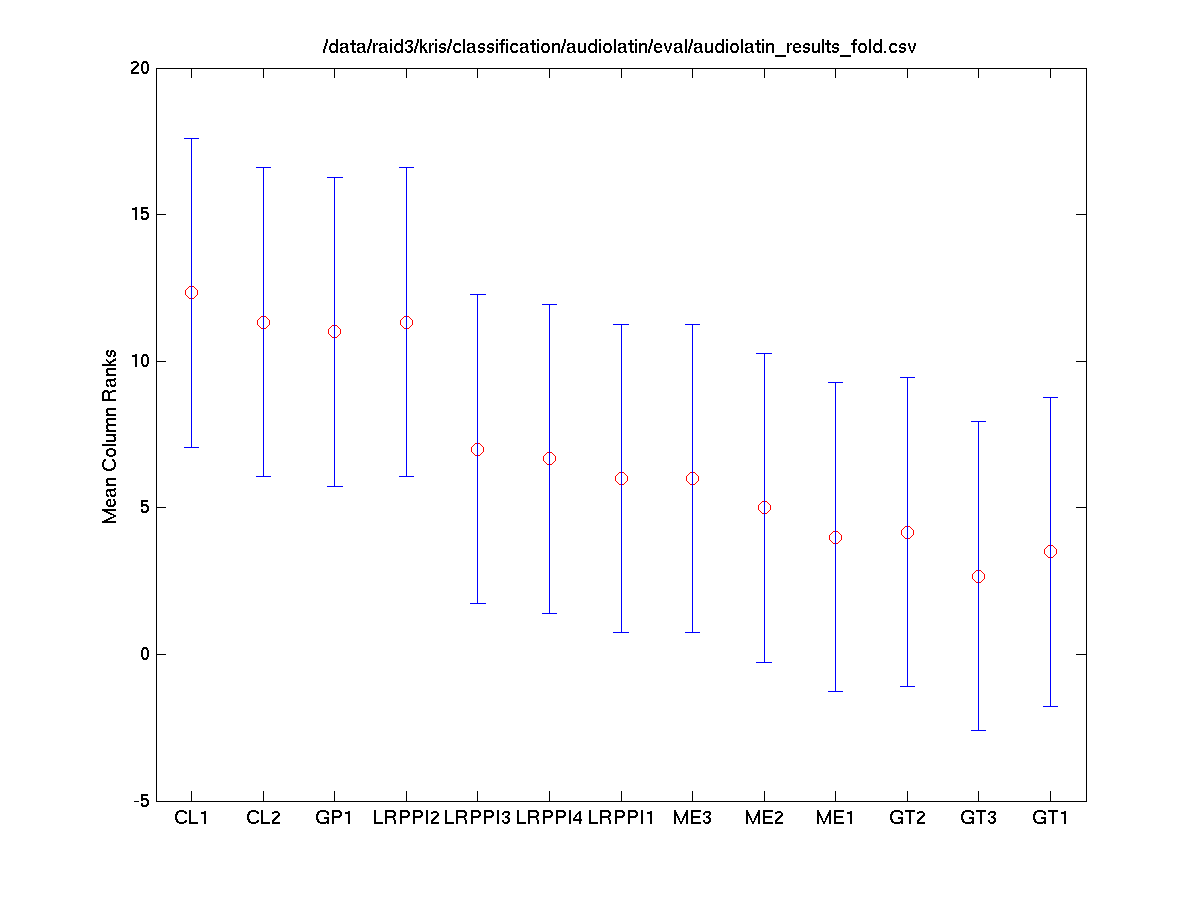
Task 1 (Mixed) Folds vs. Systems
The Friedman test was run in MATLAB against the accuracy for each fold.
Friedman's Anova Table
| Source |
SS |
df |
MS |
Chi-sq |
Prob>Chi-sq |
| Columns |
255.333 |
10 |
25.5333 |
23.21 |
0.01 |
| Error |
74.667 |
20 |
3.7333 |
|
|
| Total |
330 |
32 |
|
|
|
download these results as csv
Tukey-Kramer HSD Multi-Comparison
The Tukey-Kramer HSD multi-comparison data below was generated using the following MATLAB instruction.
Command: [c, m, h, gnames] = multicompare(stats, 'ctype', 'tukey-kramer', 'estimate', 'friedman', 'alpha', 0.05);
| TeamID |
TeamID |
Lowerbound |
Mean |
Upperbound |
Significance |
| CL1 |
CL2 |
-7.3828 |
1.3333 |
10.0495 |
FALSE |
| CL1 |
GP1 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| CL1 |
GT1 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| CL1 |
GT2 |
-6.0495 |
2.6667 |
11.3828 |
FALSE |
| CL1 |
GT3 |
-4.0495 |
4.6667 |
13.3828 |
FALSE |
| CL1 |
LRPPI1 |
-3.7162 |
5.0000 |
13.7162 |
FALSE |
| CL1 |
LRPPI2 |
-2.3828 |
6.3333 |
15.0495 |
FALSE |
| CL1 |
LRPPI3 |
-1.7162 |
7.0000 |
15.7162 |
FALSE |
| CL1 |
LRPPI4 |
-0.3828 |
8.3333 |
17.0495 |
FALSE |
| CL1 |
ME1 |
-0.3828 |
8.3333 |
17.0495 |
FALSE |
| CL2 |
GP1 |
-8.0495 |
0.6667 |
9.3828 |
FALSE |
| CL2 |
GT1 |
-8.0495 |
0.6667 |
9.3828 |
FALSE |
| CL2 |
GT2 |
-7.3828 |
1.3333 |
10.0495 |
FALSE |
| CL2 |
GT3 |
-5.3828 |
3.3333 |
12.0495 |
FALSE |
| CL2 |
LRPPI1 |
-5.0495 |
3.6667 |
12.3828 |
FALSE |
| CL2 |
LRPPI2 |
-3.7162 |
5.0000 |
13.7162 |
FALSE |
| CL2 |
LRPPI3 |
-3.0495 |
5.6667 |
14.3828 |
FALSE |
| CL2 |
LRPPI4 |
-1.7162 |
7.0000 |
15.7162 |
FALSE |
| CL2 |
ME1 |
-1.7162 |
7.0000 |
15.7162 |
FALSE |
| GP1 |
GT1 |
-8.7162 |
0.0000 |
8.7162 |
FALSE |
| GP1 |
GT2 |
-8.0495 |
0.6667 |
9.3828 |
FALSE |
| GP1 |
GT3 |
-6.0495 |
2.6667 |
11.3828 |
FALSE |
| GP1 |
LRPPI1 |
-5.7162 |
3.0000 |
11.7162 |
FALSE |
| GP1 |
LRPPI2 |
-4.3828 |
4.3333 |
13.0495 |
FALSE |
| GP1 |
LRPPI3 |
-3.7162 |
5.0000 |
13.7162 |
FALSE |
| GP1 |
LRPPI4 |
-2.3828 |
6.3333 |
15.0495 |
FALSE |
| GP1 |
ME1 |
-2.3828 |
6.3333 |
15.0495 |
FALSE |
| GT1 |
GT2 |
-8.0495 |
0.6667 |
9.3828 |
FALSE |
| GT1 |
GT3 |
-6.0495 |
2.6667 |
11.3828 |
FALSE |
| GT1 |
LRPPI1 |
-5.7162 |
3.0000 |
11.7162 |
FALSE |
| GT1 |
LRPPI2 |
-4.3828 |
4.3333 |
13.0495 |
FALSE |
| GT1 |
LRPPI3 |
-3.7162 |
5.0000 |
13.7162 |
FALSE |
| GT1 |
LRPPI4 |
-2.3828 |
6.3333 |
15.0495 |
FALSE |
| GT1 |
ME1 |
-2.3828 |
6.3333 |
15.0495 |
FALSE |
| GT2 |
GT3 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| GT2 |
LRPPI1 |
-6.3828 |
2.3333 |
11.0495 |
FALSE |
| GT2 |
LRPPI2 |
-5.0495 |
3.6667 |
12.3828 |
FALSE |
| GT2 |
LRPPI3 |
-4.3828 |
4.3333 |
13.0495 |
FALSE |
| GT2 |
LRPPI4 |
-3.0495 |
5.6667 |
14.3828 |
FALSE |
| GT2 |
ME1 |
-3.0495 |
5.6667 |
14.3828 |
FALSE |
| GT3 |
LRPPI1 |
-8.3828 |
0.3333 |
9.0495 |
FALSE |
| GT3 |
LRPPI2 |
-7.0495 |
1.6667 |
10.3828 |
FALSE |
| GT3 |
LRPPI3 |
-6.3828 |
2.3333 |
11.0495 |
FALSE |
| GT3 |
LRPPI4 |
-5.0495 |
3.6667 |
12.3828 |
FALSE |
| GT3 |
ME1 |
-5.0495 |
3.6667 |
12.3828 |
FALSE |
| LRPPI1 |
LRPPI2 |
-7.3828 |
1.3333 |
10.0495 |
FALSE |
| LRPPI1 |
LRPPI3 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| LRPPI1 |
LRPPI4 |
-5.3828 |
3.3333 |
12.0495 |
FALSE |
| LRPPI1 |
ME1 |
-5.3828 |
3.3333 |
12.0495 |
FALSE |
| LRPPI2 |
LRPPI3 |
-8.0495 |
0.6667 |
9.3828 |
FALSE |
| LRPPI2 |
LRPPI4 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| LRPPI2 |
ME1 |
-6.7162 |
2.0000 |
10.7162 |
FALSE |
| LRPPI3 |
LRPPI4 |
-7.3828 |
1.3333 |
10.0495 |
FALSE |
| LRPPI3 |
ME1 |
-7.3828 |
1.3333 |
10.0495 |
FALSE |
| LRPPI4 |
ME1 |
-8.7162 |
0.0000 |
8.7162 |
FALSE |
download these results as csv
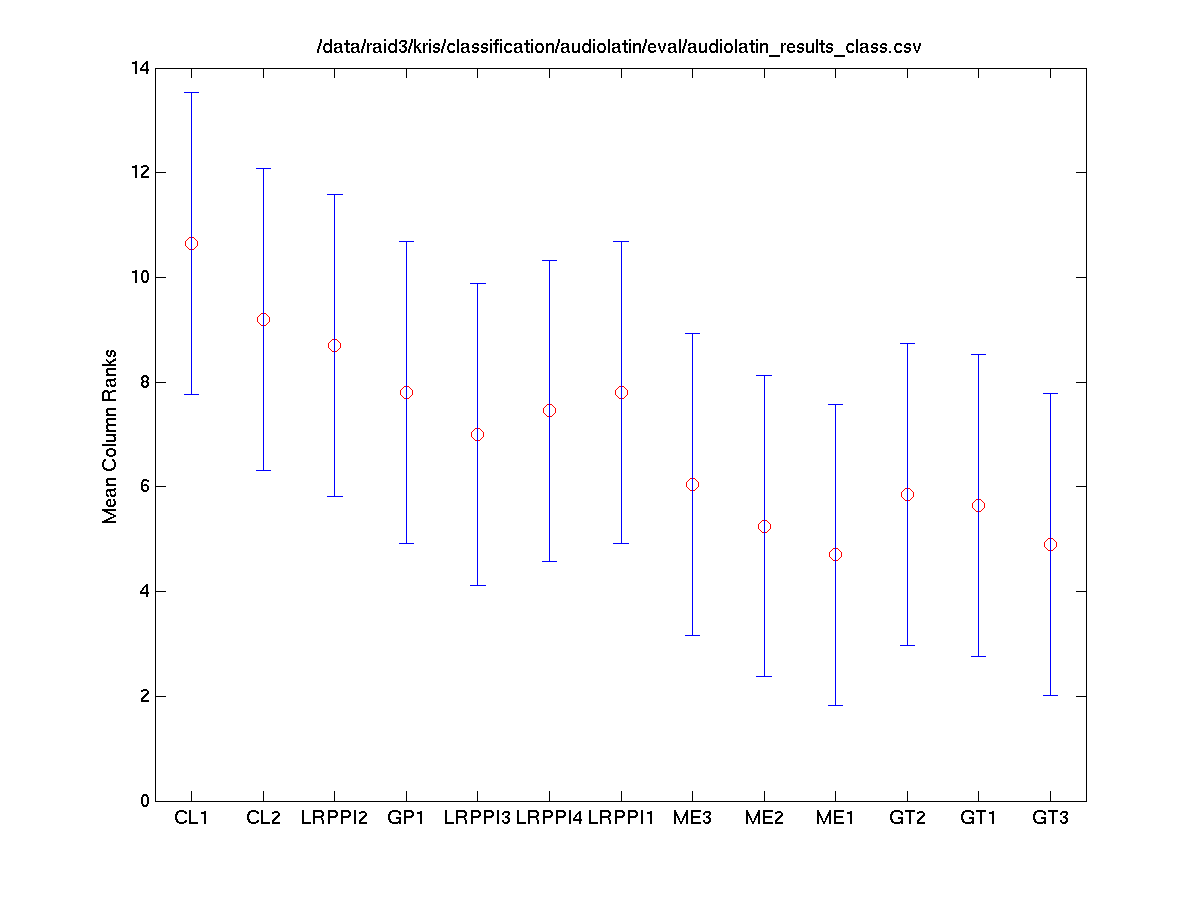
Task 2 (Latin) Classes vs. Systems
The Friedman test was run in MATLAB against the average accuracy for each class.
Friedman's Anova Table
| Source |
SS |
df |
MS |
Chi-sq |
Prob>Chi-sq |
| Columns |
235 |
10 |
23.5 |
21.38 |
0.0186 |
| Error |
864 |
90 |
9.6 |
|
|
| Total |
1099 |
109 |
|
|
|
download these results as csv
Tukey-Kramer HSD Multi-Comparison
The Tukey-Kramer HSD multi-comparison data below was generated using the following MATLAB instruction.
Command: [c, m, h, gnames] = multicompare(stats, 'ctype', 'tukey-kramer', 'estimate', 'friedman', 'alpha', 0.05);
| TeamID |
TeamID |
Lowerbound |
Mean |
Upperbound |
Significance |
| CL1 |
CL2 |
-3.7219 |
1.0500 |
5.8219 |
FALSE |
| CL1 |
GP1 |
-3.2219 |
1.5500 |
6.3219 |
FALSE |
| CL1 |
GT1 |
-2.3219 |
2.4500 |
7.2219 |
FALSE |
| CL1 |
GT2 |
-1.7219 |
3.0500 |
7.8219 |
FALSE |
| CL1 |
GT3 |
-1.7719 |
3.0000 |
7.7719 |
FALSE |
| CL1 |
LRPPI1 |
-2.1219 |
2.6500 |
7.4219 |
FALSE |
| CL1 |
LRPPI2 |
-0.2219 |
4.5500 |
9.3219 |
FALSE |
| CL1 |
LRPPI3 |
-0.7719 |
4.0000 |
8.7719 |
FALSE |
| CL1 |
LRPPI4 |
-0.6719 |
4.1000 |
8.8719 |
FALSE |
| CL1 |
ME1 |
0.1781 |
4.9500 |
9.7219 |
TRUE |
| CL2 |
GP1 |
-4.2719 |
0.5000 |
5.2719 |
FALSE |
| CL2 |
GT1 |
-3.3719 |
1.4000 |
6.1719 |
FALSE |
| CL2 |
GT2 |
-2.7719 |
2.0000 |
6.7719 |
FALSE |
| CL2 |
GT3 |
-2.8219 |
1.9500 |
6.7219 |
FALSE |
| CL2 |
LRPPI1 |
-3.1719 |
1.6000 |
6.3719 |
FALSE |
| CL2 |
LRPPI2 |
-1.2719 |
3.5000 |
8.2719 |
FALSE |
| CL2 |
LRPPI3 |
-1.8219 |
2.9500 |
7.7219 |
FALSE |
| CL2 |
LRPPI4 |
-1.7219 |
3.0500 |
7.8219 |
FALSE |
| CL2 |
ME1 |
-0.8719 |
3.9000 |
8.6719 |
FALSE |
| GP1 |
GT1 |
-3.8719 |
0.9000 |
5.6719 |
FALSE |
| GP1 |
GT2 |
-3.2719 |
1.5000 |
6.2719 |
FALSE |
| GP1 |
GT3 |
-3.3219 |
1.4500 |
6.2219 |
FALSE |
| GP1 |
LRPPI1 |
-3.6719 |
1.1000 |
5.8719 |
FALSE |
| GP1 |
LRPPI2 |
-1.7719 |
3.0000 |
7.7719 |
FALSE |
| GP1 |
LRPPI3 |
-2.3219 |
2.4500 |
7.2219 |
FALSE |
| GP1 |
LRPPI4 |
-2.2219 |
2.5500 |
7.3219 |
FALSE |
| GP1 |
ME1 |
-1.3719 |
3.4000 |
8.1719 |
FALSE |
| GT1 |
GT2 |
-4.1719 |
0.6000 |
5.3719 |
FALSE |
| GT1 |
GT3 |
-4.2219 |
0.5500 |
5.3219 |
FALSE |
| GT1 |
LRPPI1 |
-4.5719 |
0.2000 |
4.9719 |
FALSE |
| GT1 |
LRPPI2 |
-2.6719 |
2.1000 |
6.8719 |
FALSE |
| GT1 |
LRPPI3 |
-3.2219 |
1.5500 |
6.3219 |
FALSE |
| GT1 |
LRPPI4 |
-3.1219 |
1.6500 |
6.4219 |
FALSE |
| GT1 |
ME1 |
-2.2719 |
2.5000 |
7.2719 |
FALSE |
| GT2 |
GT3 |
-4.8219 |
-0.0500 |
4.7219 |
FALSE |
| GT2 |
LRPPI1 |
-5.1719 |
-0.4000 |
4.3719 |
FALSE |
| GT2 |
LRPPI2 |
-3.2719 |
1.5000 |
6.2719 |
FALSE |
| GT2 |
LRPPI3 |
-3.8219 |
0.9500 |
5.7219 |
FALSE |
| GT2 |
LRPPI4 |
-3.7219 |
1.0500 |
5.8219 |
FALSE |
| GT2 |
ME1 |
-2.8719 |
1.9000 |
6.6719 |
FALSE |
| GT3 |
LRPPI1 |
-5.1219 |
-0.3500 |
4.4219 |
FALSE |
| GT3 |
LRPPI2 |
-3.2219 |
1.5500 |
6.3219 |
FALSE |
| GT3 |
LRPPI3 |
-3.7719 |
1.0000 |
5.7719 |
FALSE |
| GT3 |
LRPPI4 |
-3.6719 |
1.1000 |
5.8719 |
FALSE |
| GT3 |
ME1 |
-2.8219 |
1.9500 |
6.7219 |
FALSE |
| LRPPI1 |
LRPPI2 |
-2.8719 |
1.9000 |
6.6719 |
FALSE |
| LRPPI1 |
LRPPI3 |
-3.4219 |
1.3500 |
6.1219 |
FALSE |
| LRPPI1 |
LRPPI4 |
-3.3219 |
1.4500 |
6.2219 |
FALSE |
| LRPPI1 |
ME1 |
-2.4719 |
2.3000 |
7.0719 |
FALSE |
| LRPPI2 |
LRPPI3 |
-5.3219 |
-0.5500 |
4.2219 |
FALSE |
| LRPPI2 |
LRPPI4 |
-5.2219 |
-0.4500 |
4.3219 |
FALSE |
| LRPPI2 |
ME1 |
-4.3719 |
0.4000 |
5.1719 |
FALSE |
| LRPPI3 |
LRPPI4 |
-4.6719 |
0.1000 |
4.8719 |
FALSE |
| LRPPI3 |
ME1 |
-3.8219 |
0.9500 |
5.7219 |
FALSE |
| LRPPI4 |
ME1 |
-3.9219 |
0.8500 |
5.6219 |
FALSE |
download these results as csv
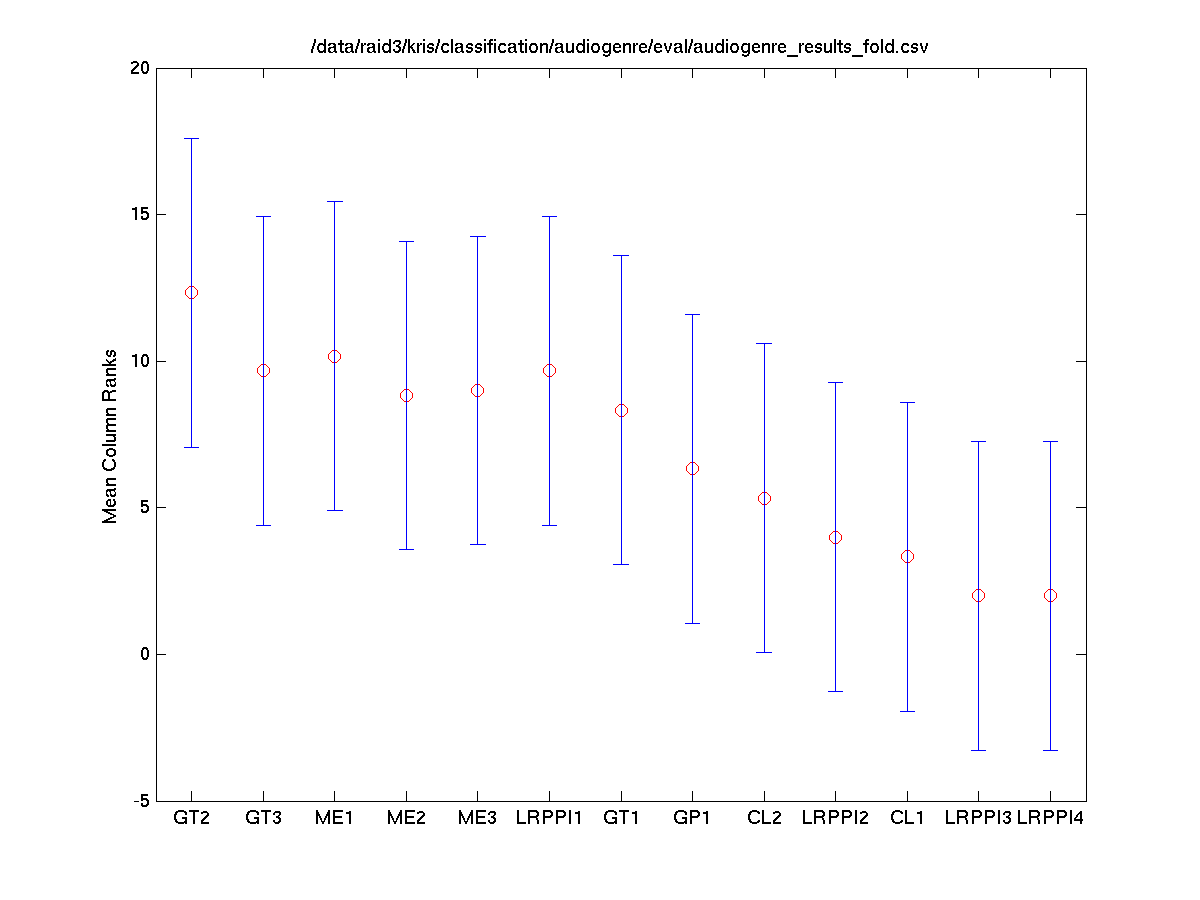
Task 2 (Latin) Folds vs. Systems
The Friedman test was run in MATLAB against the accuracy for each fold.
Friedman's Anova Table
| Source |
SS |
df |
MS |
Chi-sq |
Prob>Chi-sq |
| Columns |
265.833 |
10 |
26.5833 |
24.2 |
0.0071 |
| Error |
63.667 |
20 |
3.1833 |
|
|
| Total |
329.5 |
32 |
|
|
|
download these results as csv
Tukey-Kramer HSD Multi-Comparison
The Tukey-Kramer HSD multi-comparison data below was generated using the following MATLAB instruction.
Command: [c, m, h, gnames] = multicompare(stats, 'ctype', 'tukey-kramer', 'estimate', 'friedman', 'alpha', 0.05);
| TeamID |
TeamID |
Lowerbound |
Mean |
Upperbound |
Significance |
| CL1 |
CL2 |
-7.7095 |
1.0000 |
9.7095 |
FALSE |
| CL1 |
GP1 |
-7.3762 |
1.3333 |
10.0429 |
FALSE |
| CL1 |
GT1 |
-7.7095 |
1.0000 |
9.7095 |
FALSE |
| CL1 |
GT2 |
-4.0429 |
4.6667 |
13.3762 |
FALSE |
| CL1 |
GT3 |
-3.7095 |
5.0000 |
13.7095 |
FALSE |
| CL1 |
LRPPI1 |
-3.0429 |
5.6667 |
14.3762 |
FALSE |
| CL1 |
LRPPI2 |
-2.3762 |
6.3333 |
15.0429 |
FALSE |
| CL1 |
LRPPI3 |
-1.8762 |
6.8333 |
15.5429 |
FALSE |
| CL1 |
LRPPI4 |
-0.3762 |
8.3333 |
17.0429 |
FALSE |
| CL1 |
ME1 |
-1.2095 |
7.5000 |
16.2095 |
FALSE |
| CL2 |
GP1 |
-8.3762 |
0.3333 |
9.0429 |
FALSE |
| CL2 |
GT1 |
-8.7095 |
0.0000 |
8.7095 |
FALSE |
| CL2 |
GT2 |
-5.0429 |
3.6667 |
12.3762 |
FALSE |
| CL2 |
GT3 |
-4.7095 |
4.0000 |
12.7095 |
FALSE |
| CL2 |
LRPPI1 |
-4.0429 |
4.6667 |
13.3762 |
FALSE |
| CL2 |
LRPPI2 |
-3.3762 |
5.3333 |
14.0429 |
FALSE |
| CL2 |
LRPPI3 |
-2.8762 |
5.8333 |
14.5429 |
FALSE |
| CL2 |
LRPPI4 |
-1.3762 |
7.3333 |
16.0429 |
FALSE |
| CL2 |
ME1 |
-2.2095 |
6.5000 |
15.2095 |
FALSE |
| GP1 |
GT1 |
-9.0429 |
-0.3333 |
8.3762 |
FALSE |
| GP1 |
GT2 |
-5.3762 |
3.3333 |
12.0429 |
FALSE |
| GP1 |
GT3 |
-5.0429 |
3.6667 |
12.3762 |
FALSE |
| GP1 |
LRPPI1 |
-4.3762 |
4.3333 |
13.0429 |
FALSE |
| GP1 |
LRPPI2 |
-3.7095 |
5.0000 |
13.7095 |
FALSE |
| GP1 |
LRPPI3 |
-3.2095 |
5.5000 |
14.2095 |
FALSE |
| GP1 |
LRPPI4 |
-1.7095 |
7.0000 |
15.7095 |
FALSE |
| GP1 |
ME1 |
-2.5429 |
6.1667 |
14.8762 |
FALSE |
| GT1 |
GT2 |
-5.0429 |
3.6667 |
12.3762 |
FALSE |
| GT1 |
GT3 |
-4.7095 |
4.0000 |
12.7095 |
FALSE |
| GT1 |
LRPPI1 |
-4.0429 |
4.6667 |
13.3762 |
FALSE |
| GT1 |
LRPPI2 |
-3.3762 |
5.3333 |
14.0429 |
FALSE |
| GT1 |
LRPPI3 |
-2.8762 |
5.8333 |
14.5429 |
FALSE |
| GT1 |
LRPPI4 |
-1.3762 |
7.3333 |
16.0429 |
FALSE |
| GT1 |
ME1 |
-2.2095 |
6.5000 |
15.2095 |
FALSE |
| GT2 |
GT3 |
-8.3762 |
0.3333 |
9.0429 |
FALSE |
| GT2 |
LRPPI1 |
-7.7095 |
1.0000 |
9.7095 |
FALSE |
| GT2 |
LRPPI2 |
-7.0429 |
1.6667 |
10.3762 |
FALSE |
| GT2 |
LRPPI3 |
-6.5429 |
2.1667 |
10.8762 |
FALSE |
| GT2 |
LRPPI4 |
-5.0429 |
3.6667 |
12.3762 |
FALSE |
| GT2 |
ME1 |
-5.8762 |
2.8333 |
11.5429 |
FALSE |
| GT3 |
LRPPI1 |
-8.0429 |
0.6667 |
9.3762 |
FALSE |
| GT3 |
LRPPI2 |
-7.3762 |
1.3333 |
10.0429 |
FALSE |
| GT3 |
LRPPI3 |
-6.8762 |
1.8333 |
10.5429 |
FALSE |
| GT3 |
LRPPI4 |
-5.3762 |
3.3333 |
12.0429 |
FALSE |
| GT3 |
ME1 |
-6.2095 |
2.5000 |
11.2095 |
FALSE |
| LRPPI1 |
LRPPI2 |
-8.0429 |
0.6667 |
9.3762 |
FALSE |
| LRPPI1 |
LRPPI3 |
-7.5429 |
1.1667 |
9.8762 |
FALSE |
| LRPPI1 |
LRPPI4 |
-6.0429 |
2.6667 |
11.3762 |
FALSE |
| LRPPI1 |
ME1 |
-6.8762 |
1.8333 |
10.5429 |
FALSE |
| LRPPI2 |
LRPPI3 |
-8.2095 |
0.5000 |
9.2095 |
FALSE |
| LRPPI2 |
LRPPI4 |
-6.7095 |
2.0000 |
10.7095 |
FALSE |
| LRPPI2 |
ME1 |
-7.5429 |
1.1667 |
9.8762 |
FALSE |
| LRPPI3 |
LRPPI4 |
-7.2095 |
1.5000 |
10.2095 |
FALSE |
| LRPPI3 |
ME1 |
-8.0429 |
0.6667 |
9.3762 |
FALSE |
| LRPPI4 |
ME1 |
-9.5429 |
-0.8333 |
7.8762 |
FALSE |
download these results as csv