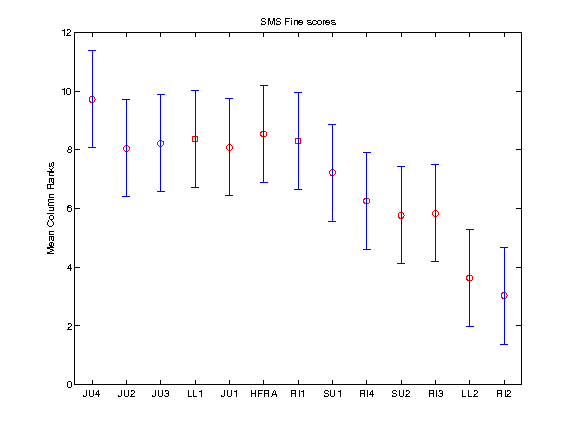
2010:Symbolic Melodic Similarity Results
Introduction
These are the results for the 2010 running of the Symbolic Melodic Similarity task set. For background information about this task set please refer to the 2010:Symbolic Melodic Similarity page.
Each system was given a query and returned the 10 most melodically similar songs from those taken from the Essen Collection (5274 pieces in the MIDI format; see ESAC Data Homepage for more information). For each query, we made four classes of error-mutations, thus the set comprises the following query classes:
- 0. No errors
- 1. One note deleted
- 2. One note inserted
- 3. One interval enlarged
- 4. One interval compressed
For each query (and its 4 mutations), the returned results (candidates) from all systems were then grouped together (query set) for evaluation by the human graders. The graders were provide with only heard perfect version against which to evaluate the candidates and did not know whether the candidates came from a perfect or mutated query. Each query/candidate set was evaluated by 1 individual grader. Using the Evalutron 6000 system, the graders gave each query/candidate pair two types of scores. Graders were asked to provide 1 categorical score with 3 categories: NS,SS,VS as explained below, and one fine score (in the range from 0 to 100).
Evalutron 6000 Summary Data
Number of evaluators = 6
Number of evaluations per query/candidate pair = 1
Number of queries per grader = 1
Total number of candidates returned = 3900
Total number of unique query/candidate pairs graded = 895
Average number of query/candidate pairs evaluated per grader: 149
Number of queries = 6 (perfect) with each perfect query error-mutated 4 different ways = 30
General Legend
General Legend
| Sub code | Submission name | Abstract | Contributors |
|---|---|---|---|
| HFRA1 | SMS simbals | Pierre Hanna, Pascal Ferraro, Matthias Robine, Julien Allali | |
| JU1 | SMS-Domain | Julián Urbano, Juan Lloréns, Jorge Morato, Sonia Sánchez-Cuadrado | |
| JU2 | SMS-PitchDeriv | Julián Urbano, Juan Lloréns, Jorge Morato, Sonia Sánchez-Cuadrado | |
| JU3 | SMS-ParamDeriv | Julián Urbano, Juan Lloréns, Jorge Morato, Sonia Sánchez-Cuadrado | |
| JU4 | SMS-Shape | Julián Urbano, Juan Lloréns, Jorge Morato, Sonia Sánchez-Cuadrado | |
| LL1 | CbrahmsS2 | Mika Laitinen, Kjell Lemström | |
| LL2 | CbrahmsW2 | Mika Laitinen, Kjell Lemström | |
| RI1 | UAC | David Rizo, José Manuel Iñesta | |
| RI2 | UAT | David Rizo, José Manuel Iñesta | |
| RI3 | UAT3 | David Rizo, José Manuel Iñesta | |
| RI4 | UAPR | David Rizo, José Manuel Iñesta | |
| SU1 | NGR5 | Iman Suyoto, Alexandra Uitdenbogerd | |
| SU2 | PIOI | Iman Suyoto, Alexandra Uitdenbogerd |
Broad Categories
NS = Not Similar
SS = Somewhat Similar
VS = Very Similar
Table Headings
ADR = Average Dynamic Recall
NRGB = Normalize Recall at Group Boundaries
AP = Average Precision (non-interpolated)
PND = Precision at N Documents
Calculating Summary Measures
Fine(1) = Sum of fine-grained human similarity decisions (0-100).
PSum(1) = Sum of human broad similarity decisions: NS=0, SS=1, VS=2.
WCsum(1) = 'World Cup' scoring: NS=0, SS=1, VS=3 (rewards Very Similar).
SDsum(1) = 'Stephen Downie' scoring: NS=0, SS=1, VS=4 (strongly rewards Very Similar).
Greater0(1) = NS=0, SS=1, VS=1 (binary relevance judgment).
Greater1(1) = NS=0, SS=0, VS=1 (binary relevance judgment using only Very Similar).
(1)Normalized to the range 0 to 1.
Summary Results
Run Times
| Participant | Runtime (min) | Machine |
|---|---|---|
| HFRA1 | 16 | nema-c-1-2 |
| JU1 | 7 | win-0-1-1 |
| JU2 | 15 | win-0-1-1 |
| JU3 | 21 | win-0-1-1 |
| JU4 | 20 | win-0-1-1 |
| LL1 | 205 | compute-1-0 |
| LL2 | 286 | compute-1-0 |
| RI1 | 13 | nema-c-1-2 |
| RI2 | 4 | nema-c-1-2 |
| RI3 | 6 | nema-c-1-2 |
| RI4 | 5 | nema-c-1-2 |
| SU1 | 26 | compute-1-0 |
| SU2 | 8 | compute-1-0 |
Overall Scores (Includes Perfect and Error Candidates)
| Overall | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2472 | 0.3069 | 0.3091 | 0.3168 | 0.3713 | 0.3077 | 0.2105 | 0.2744 | 0.1078 | 0.2234 | 0.1384 | 0.2904 | 0.2487 |
| NRGB | 0.2405 | 0.2974 | 0.2941 | 0.2877 | 0.3278 | 0.2989 | 0.1730 | 0.2751 | 0.1167 | 0.2028 | 0.1581 | 0.2651 | 0.2361 |
| AP | 0.1914 | 0.2998 | 0.2987 | 0.3012 | 0.3492 | 0.2398 | 0.1816 | 0.2560 | 0.0660 | 0.1448 | 0.1281 | 0.2454 | 0.2265 |
| PND | 0.2829 | 0.3726 | 0.3726 | 0.3681 | 0.3985 | 0.3410 | 0.2226 | 0.3530 | 0.1390 | 0.2241 | 0.2062 | 0.3353 | 0.2839 |
| Fine | 0.5625 | 0.5793 | 0.5832 | 0.5806 | 0.6055 | 0.5799 | 0.4140 | 0.5453 | 0.3794 | 0.4705 | 0.5219 | 0.5356 | 0.5099 |
| Psum | 0.5783 | 0.6133 | 0.6200 | 0.6150 | 0.6417 | 0.6000 | 0.4017 | 0.5817 | 0.3583 | 0.4983 | 0.5583 | 0.5433 | 0.5317 |
| WCsum | 0.5122 | 0.5589 | 0.5633 | 0.5589 | 0.5800 | 0.5478 | 0.3489 | 0.5200 | 0.3022 | 0.4289 | 0.4789 | 0.5033 | 0.4844 |
| SDsum | 0.4792 | 0.5317 | 0.5350 | 0.5308 | 0.5492 | 0.5217 | 0.3225 | 0.4892 | 0.2742 | 0.3942 | 0.4392 | 0.4833 | 0.4608 |
| Greater0 | 0.7767 | 0.7767 | 0.7900 | 0.7833 | 0.8267 | 0.7567 | 0.5600 | 0.7667 | 0.5267 | 0.7067 | 0.7967 | 0.6633 | 0.6733 |
| Greater1 | 0.3800 | 0.4500 | 0.4500 | 0.4467 | 0.4567 | 0.4433 | 0.2433 | 0.3967 | 0.1900 | 0.2900 | 0.3200 | 0.4233 | 0.3900 |
Scores by Query Error Types
| No_Errors | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2576 | 0.3492 | 0.3492 | 0.3585 | 0.3946 | 0.3213 | 0.1688 | 0.2799 | 0.1536 | 0.3026 | 0.1346 | 0.2811 | 0.2822 |
| NRGB | 0.2420 | 0.3279 | 0.3279 | 0.3279 | 0.3445 | 0.2973 | 0.1428 | 0.2903 | 0.1640 | 0.2688 | 0.1615 | 0.2543 | 0.2388 |
| AP | 0.2122 | 0.3470 | 0.3512 | 0.3574 | 0.3898 | 0.2708 | 0.1823 | 0.2634 | 0.1107 | 0.2279 | 0.1461 | 0.2457 | 0.2612 |
| PND | 0.2750 | 0.4097 | 0.4097 | 0.4097 | 0.4264 | 0.3847 | 0.2250 | 0.3556 | 0.2014 | 0.2847 | 0.2000 | 0.3306 | 0.2750 |
| Fine | 0.5452 | 0.6293 | 0.6227 | 0.6155 | 0.6413 | 0.5958 | 0.4188 | 0.5450 | 0.4162 | 0.5172 | 0.5545 | 0.5755 | 0.5328 |
| Psum | 0.5667 | 0.6667 | 0.6583 | 0.6500 | 0.6750 | 0.6250 | 0.4083 | 0.5750 | 0.4167 | 0.5500 | 0.6000 | 0.5833 | 0.5583 |
| WCsum | 0.5000 | 0.6056 | 0.6000 | 0.5944 | 0.6111 | 0.5722 | 0.3611 | 0.5167 | 0.3667 | 0.4833 | 0.5167 | 0.5389 | 0.5111 |
| SDsum | 0.4667 | 0.5750 | 0.5708 | 0.5667 | 0.5792 | 0.5458 | 0.3375 | 0.4875 | 0.3417 | 0.4500 | 0.4750 | 0.5167 | 0.4875 |
| Greater0 | 0.7667 | 0.8500 | 0.8333 | 0.8167 | 0.8667 | 0.7833 | 0.5500 | 0.7500 | 0.5667 | 0.7500 | 0.8500 | 0.7167 | 0.7000 |
| Greater1 | 0.3667 | 0.4833 | 0.4833 | 0.4833 | 0.4833 | 0.4667 | 0.2667 | 0.4000 | 0.2667 | 0.3500 | 0.3500 | 0.4500 | 0.4167 |
| Deleted | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2440 | 0.3305 | 0.3305 | 0.3691 | 0.4068 | 0.2773 | 0.2174 | 0.2745 | 0.0329 | 0.1251 | 0.1396 | 0.3135 | 0.2400 |
| NRGB | 0.2423 | 0.3313 | 0.3313 | 0.3299 | 0.3577 | 0.2450 | 0.1855 | 0.2487 | 0.0567 | 0.1200 | 0.1443 | 0.2973 | 0.2362 |
| AP | 0.2339 | 0.3547 | 0.3547 | 0.3741 | 0.4036 | 0.2194 | 0.2305 | 0.2555 | 0.0095 | 0.0542 | 0.0805 | 0.2808 | 0.2177 |
| PND | 0.3241 | 0.4278 | 0.4278 | 0.4278 | 0.4444 | 0.2889 | 0.2370 | 0.3222 | 0.0500 | 0.1333 | 0.1852 | 0.3870 | 0.2704 |
| Fine | 0.5533 | 0.6165 | 0.6165 | 0.6198 | 0.6323 | 0.5487 | 0.4182 | 0.5242 | 0.2983 | 0.3795 | 0.4598 | 0.5328 | 0.5168 |
| Psum | 0.5667 | 0.6583 | 0.6583 | 0.6667 | 0.6750 | 0.5500 | 0.4167 | 0.5500 | 0.2333 | 0.3667 | 0.4833 | 0.5417 | 0.5167 |
| WCsum | 0.5111 | 0.6056 | 0.6056 | 0.6111 | 0.6167 | 0.5056 | 0.3611 | 0.5000 | 0.1833 | 0.3056 | 0.4167 | 0.5056 | 0.4722 |
| SDsum | 0.4833 | 0.5792 | 0.5792 | 0.5833 | 0.5875 | 0.4833 | 0.3333 | 0.4750 | 0.1583 | 0.2750 | 0.3833 | 0.4875 | 0.4500 |
| Greater0 | 0.7333 | 0.8167 | 0.8167 | 0.8333 | 0.8500 | 0.6833 | 0.5833 | 0.7000 | 0.3833 | 0.5500 | 0.6833 | 0.6500 | 0.6500 |
| Greater1 | 0.4000 | 0.5000 | 0.5000 | 0.5000 | 0.5000 | 0.4167 | 0.2500 | 0.4000 | 0.0833 | 0.1833 | 0.2833 | 0.4333 | 0.3833 |
| Inserted | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2324 | 0.2751 | 0.3156 | 0.3193 | 0.3555 | 0.3131 | 0.3225 | 0.2516 | 0.0488 | 0.1556 | 0.1487 | 0.2978 | 0.2253 |
| NRGB | 0.2253 | 0.2568 | 0.2635 | 0.2735 | 0.3119 | 0.3202 | 0.2693 | 0.2354 | 0.0530 | 0.1406 | 0.1598 | 0.2671 | 0.2303 |
| AP | 0.1719 | 0.2679 | 0.2899 | 0.2803 | 0.3217 | 0.2098 | 0.2216 | 0.2209 | 0.0218 | 0.0963 | 0.1139 | 0.2360 | 0.1886 |
| PND | 0.2656 | 0.3587 | 0.3754 | 0.3587 | 0.3921 | 0.2899 | 0.2751 | 0.3288 | 0.0685 | 0.1942 | 0.2146 | 0.3365 | 0.3198 |
| Fine | 0.5512 | 0.5880 | 0.6032 | 0.5932 | 0.6205 | 0.5545 | 0.4202 | 0.5315 | 0.3300 | 0.4363 | 0.5157 | 0.5088 | 0.5070 |
| Psum | 0.5500 | 0.6417 | 0.6583 | 0.6417 | 0.6750 | 0.5667 | 0.4083 | 0.5750 | 0.3083 | 0.4750 | 0.5500 | 0.5167 | 0.5500 |
| WCsum | 0.4889 | 0.5778 | 0.5944 | 0.5778 | 0.6056 | 0.5111 | 0.3556 | 0.5000 | 0.2389 | 0.4000 | 0.4667 | 0.4778 | 0.4944 |
| SDsum | 0.4583 | 0.5458 | 0.5625 | 0.5458 | 0.5708 | 0.4833 | 0.3292 | 0.4625 | 0.2042 | 0.3625 | 0.4250 | 0.4583 | 0.4667 |
| Greater0 | 0.7333 | 0.8333 | 0.8500 | 0.8333 | 0.8833 | 0.7333 | 0.5667 | 0.8000 | 0.5167 | 0.7000 | 0.8000 | 0.6333 | 0.7167 |
| Greater1 | 0.3667 | 0.4500 | 0.4667 | 0.4500 | 0.4667 | 0.4000 | 0.2500 | 0.3500 | 0.1000 | 0.2500 | 0.3000 | 0.4000 | 0.3833 |
| Enlarged | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2349 | 0.2665 | 0.2609 | 0.2352 | 0.3300 | 0.3171 | 0.1763 | 0.3018 | 0.1346 | 0.2593 | 0.1379 | 0.2691 | 0.2548 |
| NRGB | 0.2289 | 0.2804 | 0.2637 | 0.2350 | 0.2961 | 0.3428 | 0.1381 | 0.3171 | 0.1403 | 0.2394 | 0.1670 | 0.2514 | 0.2489 |
| AP | 0.1564 | 0.2257 | 0.2095 | 0.1907 | 0.2693 | 0.2563 | 0.1179 | 0.2733 | 0.0730 | 0.1450 | 0.1618 | 0.2106 | 0.2315 |
| PND | 0.2389 | 0.3111 | 0.2944 | 0.2889 | 0.3389 | 0.3889 | 0.1778 | 0.3833 | 0.1444 | 0.2444 | 0.2444 | 0.2944 | 0.2667 |
| Fine | 0.5760 | 0.5027 | 0.4973 | 0.5047 | 0.5455 | 0.6010 | 0.3932 | 0.5692 | 0.4278 | 0.5153 | 0.5333 | 0.5248 | 0.4858 |
| Psum | 0.6000 | 0.5167 | 0.5083 | 0.5250 | 0.5667 | 0.6417 | 0.3667 | 0.5917 | 0.4167 | 0.5833 | 0.5750 | 0.5250 | 0.5000 |
| WCsum | 0.5222 | 0.4722 | 0.4611 | 0.4722 | 0.5056 | 0.5944 | 0.3167 | 0.5333 | 0.3556 | 0.5056 | 0.4944 | 0.4833 | 0.4556 |
| SDsum | 0.4833 | 0.4500 | 0.4375 | 0.4458 | 0.4750 | 0.5708 | 0.2917 | 0.5042 | 0.3250 | 0.4667 | 0.4542 | 0.4625 | 0.4333 |
| Greater0 | 0.8333 | 0.6500 | 0.6500 | 0.6833 | 0.7500 | 0.7833 | 0.5167 | 0.7667 | 0.6000 | 0.8167 | 0.8167 | 0.6500 | 0.6333 |
| Greater1 | 0.3667 | 0.3833 | 0.3667 | 0.3667 | 0.3833 | 0.5000 | 0.2167 | 0.4167 | 0.2333 | 0.3500 | 0.3333 | 0.4000 | 0.3667 |
| Compressed | HFRA | JU1 | JU2 | JU3 | JU4 | LL1 | LL2 | RI1 | RI2 | RI3 | RI4 | SU1 | SU2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADR | 0.2671 | 0.3132 | 0.2891 | 0.3017 | 0.3694 | 0.3096 | 0.1673 | 0.2643 | 0.1691 | 0.2745 | 0.1313 | 0.2903 | 0.2413 |
| NRGB | 0.2642 | 0.2908 | 0.2842 | 0.2723 | 0.3288 | 0.2891 | 0.1293 | 0.2838 | 0.1697 | 0.2451 | 0.1580 | 0.2552 | 0.2264 |
| AP | 0.1828 | 0.3036 | 0.2884 | 0.3036 | 0.3617 | 0.2430 | 0.1555 | 0.2670 | 0.1149 | 0.2009 | 0.1383 | 0.2540 | 0.2334 |
| PND | 0.3110 | 0.3555 | 0.3555 | 0.3555 | 0.3907 | 0.3527 | 0.1983 | 0.3751 | 0.2305 | 0.2638 | 0.1870 | 0.3282 | 0.2877 |
| Fine | 0.5868 | 0.5602 | 0.5762 | 0.5700 | 0.5877 | 0.5997 | 0.4197 | 0.5567 | 0.4247 | 0.5040 | 0.5462 | 0.5360 | 0.5068 |
| Psum | 0.6083 | 0.5833 | 0.6167 | 0.5917 | 0.6167 | 0.6167 | 0.4083 | 0.6167 | 0.4167 | 0.5167 | 0.5833 | 0.5500 | 0.5333 |
| WCsum | 0.5389 | 0.5333 | 0.5556 | 0.5389 | 0.5611 | 0.5556 | 0.3500 | 0.5500 | 0.3667 | 0.4500 | 0.5000 | 0.5111 | 0.4889 |
| SDsum | 0.5042 | 0.5083 | 0.5250 | 0.5125 | 0.5333 | 0.5250 | 0.3208 | 0.5167 | 0.3417 | 0.4167 | 0.4583 | 0.4917 | 0.4667 |
| Greater0 | 0.8167 | 0.7333 | 0.8000 | 0.7500 | 0.7833 | 0.8000 | 0.5833 | 0.8167 | 0.5667 | 0.7167 | 0.8333 | 0.6667 | 0.6667 |
| Greater1 | 0.4000 | 0.4333 | 0.4333 | 0.4333 | 0.4500 | 0.4333 | 0.2333 | 0.4167 | 0.2667 | 0.3167 | 0.3333 | 0.4333 | 0.4000 |
Friedman Test with Multiple Comparisons Results (p=0.05)
The Friedman test was run in MATLAB against the Fine summary data over the 30 queries.
Command: [c,m,h,gnames] = multcompare(stats, 'ctype', 'tukey-kramer','estimate', 'friedman', 'alpha', 0.05);
file /nema-raid/www/mirex/results/2010/sms/sum_friedman_fine.csv not found
| TeamID | TeamID | Lowerbound | Mean | Upperbound | Significance |
|---|---|---|---|---|---|
| JU4 | JU2 | -1.6410 | 1.6667 | 4.9743 | FALSE |
| JU4 | JU3 | -1.8077 | 1.5000 | 4.8077 | FALSE |
| JU4 | LL1 | -1.9577 | 1.3500 | 4.6577 | FALSE |
| JU4 | JU1 | -1.6743 | 1.6333 | 4.9410 | FALSE |
| JU4 | HFRA | -2.1243 | 1.1833 | 4.4910 | FALSE |
| JU4 | RI1 | -1.8910 | 1.4167 | 4.7243 | FALSE |
| JU4 | SU1 | -0.8077 | 2.5000 | 5.8077 | FALSE |
| JU4 | RI4 | 0.1590 | 3.4667 | 6.7743 | TRUE |
| JU4 | SU2 | 0.6423 | 3.9500 | 7.2577 | TRUE |
| JU4 | RI3 | 0.5757 | 3.8833 | 7.1910 | TRUE |
| JU4 | LL2 | 2.7757 | 6.0833 | 9.3910 | TRUE |
| JU4 | RI2 | 3.3757 | 6.6833 | 9.9910 | TRUE |
| JU2 | JU3 | -3.4743 | -0.1667 | 3.1410 | FALSE |
| JU2 | LL1 | -3.6243 | -0.3167 | 2.9910 | FALSE |
| JU2 | JU1 | -3.3410 | -0.0333 | 3.2743 | FALSE |
| JU2 | HFRA | -3.7910 | -0.4833 | 2.8243 | FALSE |
| JU2 | RI1 | -3.5577 | -0.2500 | 3.0577 | FALSE |
| JU2 | SU1 | -2.4743 | 0.8333 | 4.1410 | FALSE |
| JU2 | RI4 | -1.5077 | 1.8000 | 5.1077 | FALSE |
| JU2 | SU2 | -1.0243 | 2.2833 | 5.5910 | FALSE |
| JU2 | RI3 | -1.0910 | 2.2167 | 5.5243 | FALSE |
| JU2 | LL2 | 1.1090 | 4.4167 | 7.7243 | TRUE |
| JU2 | RI2 | 1.7090 | 5.0167 | 8.3243 | TRUE |
| JU3 | LL1 | -3.4577 | -0.1500 | 3.1577 | FALSE |
| JU3 | JU1 | -3.1743 | 0.1333 | 3.4410 | FALSE |
| JU3 | HFRA | -3.6243 | -0.3167 | 2.9910 | FALSE |
| JU3 | RI1 | -3.3910 | -0.0833 | 3.2243 | FALSE |
| JU3 | SU1 | -2.3077 | 1.0000 | 4.3077 | FALSE |
| JU3 | RI4 | -1.3410 | 1.9667 | 5.2743 | FALSE |
| JU3 | SU2 | -0.8577 | 2.4500 | 5.7577 | FALSE |
| JU3 | RI3 | -0.9243 | 2.3833 | 5.6910 | FALSE |
| JU3 | LL2 | 1.2757 | 4.5833 | 7.8910 | TRUE |
| JU3 | RI2 | 1.8757 | 5.1833 | 8.4910 | TRUE |
| LL1 | JU1 | -3.0243 | 0.2833 | 3.5910 | FALSE |
| LL1 | HFRA | -3.4743 | -0.1667 | 3.1410 | FALSE |
| LL1 | RI1 | -3.2410 | 0.0667 | 3.3743 | FALSE |
| LL1 | SU1 | -2.1577 | 1.1500 | 4.4577 | FALSE |
| LL1 | RI4 | -1.1910 | 2.1167 | 5.4243 | FALSE |
| LL1 | SU2 | -0.7077 | 2.6000 | 5.9077 | FALSE |
| LL1 | RI3 | -0.7743 | 2.5333 | 5.8410 | FALSE |
| LL1 | LL2 | 1.4257 | 4.7333 | 8.0410 | TRUE |
| LL1 | RI2 | 2.0257 | 5.3333 | 8.6410 | TRUE |
| JU1 | HFRA | -3.7577 | -0.4500 | 2.8577 | FALSE |
| JU1 | RI1 | -3.5243 | -0.2167 | 3.0910 | FALSE |
| JU1 | SU1 | -2.4410 | 0.8667 | 4.1743 | FALSE |
| JU1 | RI4 | -1.4743 | 1.8333 | 5.1410 | FALSE |
| JU1 | SU2 | -0.9910 | 2.3167 | 5.6243 | FALSE |
| JU1 | RI3 | -1.0577 | 2.2500 | 5.5577 | FALSE |
| JU1 | LL2 | 1.1423 | 4.4500 | 7.7577 | TRUE |
| JU1 | RI2 | 1.7423 | 5.0500 | 8.3577 | TRUE |
| HFRA | RI1 | -3.0743 | 0.2333 | 3.5410 | FALSE |
| HFRA | SU1 | -1.9910 | 1.3167 | 4.6243 | FALSE |
| HFRA | RI4 | -1.0243 | 2.2833 | 5.5910 | FALSE |
| HFRA | SU2 | -0.5410 | 2.7667 | 6.0743 | FALSE |
| HFRA | RI3 | -0.6077 | 2.7000 | 6.0077 | FALSE |
| HFRA | LL2 | 1.5923 | 4.9000 | 8.2077 | TRUE |
| HFRA | RI2 | 2.1923 | 5.5000 | 8.8077 | TRUE |
| RI1 | SU1 | -2.2243 | 1.0833 | 4.3910 | FALSE |
| RI1 | RI4 | -1.2577 | 2.0500 | 5.3577 | FALSE |
| RI1 | SU2 | -0.7743 | 2.5333 | 5.8410 | FALSE |
| RI1 | RI3 | -0.8410 | 2.4667 | 5.7743 | FALSE |
| RI1 | LL2 | 1.3590 | 4.6667 | 7.9743 | TRUE |
| RI1 | RI2 | 1.9590 | 5.2667 | 8.5743 | TRUE |
| SU1 | RI4 | -2.3410 | 0.9667 | 4.2743 | FALSE |
| SU1 | SU2 | -1.8577 | 1.4500 | 4.7577 | FALSE |
| SU1 | RI3 | -1.9243 | 1.3833 | 4.6910 | FALSE |
| SU1 | LL2 | 0.2757 | 3.5833 | 6.8910 | TRUE |
| SU1 | RI2 | 0.8757 | 4.1833 | 7.4910 | TRUE |
| RI4 | SU2 | -2.8243 | 0.4833 | 3.7910 | FALSE |
| RI4 | RI3 | -2.8910 | 0.4167 | 3.7243 | FALSE |
| RI4 | LL2 | -0.6910 | 2.6167 | 5.9243 | FALSE |
| RI4 | RI2 | -0.0910 | 3.2167 | 6.5243 | FALSE |
| SU2 | RI3 | -3.3743 | -0.0667 | 3.2410 | FALSE |
| SU2 | LL2 | -1.1743 | 2.1333 | 5.4410 | FALSE |
| SU2 | RI2 | -0.5743 | 2.7333 | 6.0410 | FALSE |
| RI3 | LL2 | -1.1077 | 2.2000 | 5.5077 | FALSE |
| RI3 | RI2 | -0.5077 | 2.8000 | 6.1077 | FALSE |
| LL2 | RI2 | -2.7077 | 0.6000 | 3.9077 | FALSE |
Raw Scores
The raw data derived from the Evalutron 6000 human evaluations are located on the 2010:Symbolic Melodic Similarity Raw Data page.